2) Statistical analysis on a shape space¶
%matplotlib inline
import matplotlib.pyplot as plt
import plotly
# run at the start of every notebook
plotly.offline.init_notebook_mode()
# Mandatory imports...
import numpy as np
from numpy import random
import torch
from torch import tensor
from torch.nn import Parameter
# Custom modules:
from kendall_triangles import KendallTriangles # Fancy visualization
from model import Model # Optimization blackbox
from display import plot # Simple plotting routine for triangles
# Finally, emulate complex numbers with pairs of floats:
from torch_complex import rot,normalize,herm,angle,mod2,mod,comp
A) Computing a Fréchet mean¶
In the previous notebook, we've seen that the set of triangles "up to similarities" can be endowed with a canonical, spherical metric structure. The arc (or geodesic) distance between two triangle shapes is given by:
def arc( A, B ) :
"'Geodesic' distance between two triangles A and B."
a, b = normalize(A), normalize(B)
return mod( herm(a,b) ).acos()
We now implement some (simple) statistical operations in this nonlinear space. Our key message: the standard statistician's toolbox can be generalized to Riemannian manifolds, both from a theoretical and a practical point of view.
First, let us generate some arbitrary population of shapes:
N = 100
A_k = torch.stack((
.05*torch.randn(100,2)+tensor([-.5,-.5]),
.05*torch.randn(100,2)+tensor([+.5,0. ]),
.50*torch.randn(100,2)+tensor([ 0., 1.]),
), 1) + 3*torch.randn(100,1,2)
And display it in the 2D plane:
plt.figure()
plot(A_k[:20], "green")
plt.axis("equal")
plt.show()
In the shape space:
M = KendallTriangles()
M.add_markers(A_k, "green", "A_k", .5)
M.generate_spherical_sampled_data()
M.show()
M.show_glyphs()
M.show_pieces()
M.iplot('Our triangles on the Kendall sphere')
Given such an empirical distribution $A_k$, what it the simplest statistic that we could compute? The mean, obviously. But beware: on a sphere, there is no such thing as a "$+$" operator.
One of the most sensible ways of generalizing the mean to an arbitrary metric space $(X,\text{d})$ is the least-square definition. For any order $p>0$, we thus define the $p$-Fréchet mean of the $A_k$'s through:
$$ F_p(A_1,\dots,A_N) ~=~ \arg\min_{x\in X} \,\sum_{k=1}^N \text{d}(x,A_k)^p $$
Exercise 1: Show that in a Euclidean vector space, the Fréchet means of order 1 and 2 respectively coincide with the median and the arithmetic mean of the distribution.
class FrechetMean(Model) :
"Find the Frechet mean of a population of polygons."
def __init__(self, guess, p=2) :
"Defines an initial guess and the Frechet exponent."
super(Model, self).__init__()
self.x = Parameter(guess)
self.p = p
def __call__(self) :
"Returns the current guess."
return self.x
def cost(self, targets) :
"Returns the mean p-distance to the current guess."
return ( arc(targets, self())**self.p ).mean()
Using PyTorch, computing a Fréchet mean is that simple:
# Initial guess: any non-degenerate triangle should be ok
A0 = tensor([ [-1.,0.], [1.,0.], [0.,1.] ])
# Frechet means of order 1 and 2
FMedian = FrechetMean( A0, p=1 )
FMean = FrechetMean( A0, p=2 )
FMedian.fit( A_k )
FMean.fit( A_k )
# For a clean display, let's normalize everybody
median = normalize( FMedian())
mean = normalize( FMean() )
a_k = normalize( A_k )
Without aligning the shapes onto a reference mean, displaying the population is not easy:
plt.figure()
plot( a_k[:10] , "green")
plot( mean, "red")
plot( median, "blue")
plt.axis("equal")
plt.show()
Thankfully, the Kendall sphere lets us check that our computations are correct:
M = KendallTriangles()
M.add_markers(a_k, "green", "A_k", .5)
M.add_markers(mean, "red", "mean")
M.add_markers(median, "blue", "median")
M.generate_spherical_sampled_data()
M.show()
M.show_glyphs()
M.show_pieces()
M.iplot('Mean and Median of our population')
Exercise 2: What's the difference between using a mean and a median?
B) Computing a geodesic¶
Under mild technical assumptions, Riemannian manifolds let us define geodesic curves through an order 2 ODE. If $a$ and $b$ are two points in the manifold, and if $v$ is some tangent vector at location $a$, $\text{Exp}_a(v)$ will denote the position at time $t=1$ of the unique geodesic curve $\gamma_t$ such that
$$ \gamma_0 ~=~a ~~~~~\text{and}~~~~~ \dot{\gamma}_0~=~v. $$
The length of such a geodesic curve is simply equal to the norm of the tangent vector $v$ at position $a$.
A standard result of Riemannian geometry is that for any reference point $a$, such an Exponential map defines an isomorphism from a neighborhood of $0$ (on the tangent plane) to a neighborhood of $a$ (on the manifold).
We can thus define the $\text{Log}_a$ map as the inverse of $\text{Exp}_a$ in a neighborhood of $a$, and use it to represent the curved manifold in the tangent vector space:
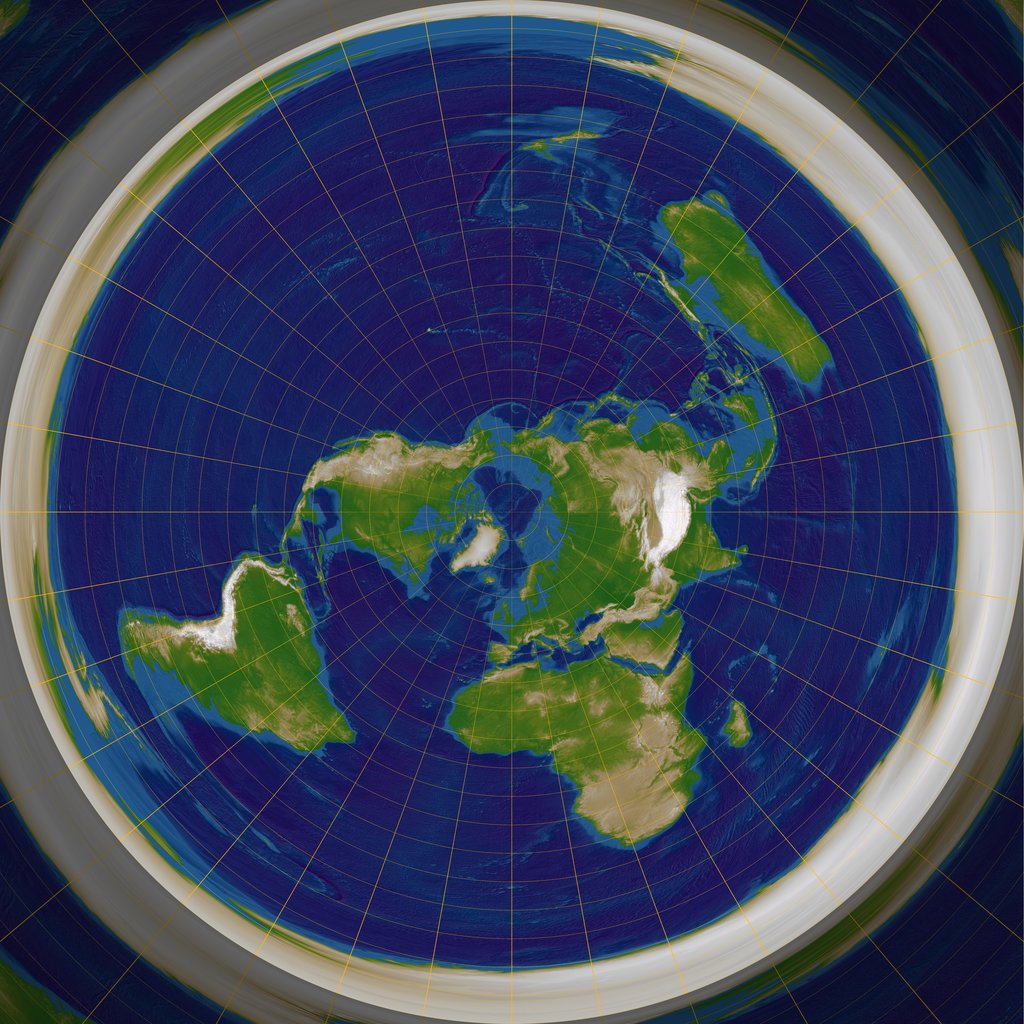 Exponential chart of the Earth, at the North pole. Image from Wikipedia, by RokerHRO.
Exponential chart of the Earth, at the North pole. Image from Wikipedia, by RokerHRO.
Most often, the $\text{Exp}$ and $\text{Log}$ operators are tedious to implement. But in the very specific (and important!) case of polygonal shapes up to similarities, the classic Kendall theory provides us with the following formulas:
def log(a,b):
"""
Assuming that a and b are both normalized, returns
the Riemannian logarithm of b at point a.
"""
# Optimally rotate b onto a
bsa = herm(b, a)
b = rot(b, angle(bsa))
# Compute the normal vector
c = b - (a*b).sum()*a
c = c / (c**2).sum().sqrt()
L = arc(a,b)
return L*c
def exp(a,v):
"Shoots along the tangent vector v at location a."
L = (v**2).sum().sqrt()
return a*torch.cos(L) + (v/L)*torch.sin(L)
def geodesic(a,b):
"""
Returns the geodesic between two *normalized* polygons a and b,
sampled on 10 timesteps with the normal tangent vector at a.
"""
v = log(a,b)
g_t = [ exp(a, t*v) \
for t in torch.linspace(0,1,11) ]
return torch.stack( g_t )
Exercise 3: Please convince yourself that it works!
# Compute an arbitrary geodesic segment
a = a_k[1]
g = geodesic(mean, a)
# Display on the phere
M = KendallTriangles()
M.add_markers(g, "green", "geodesic", .5)
M.add_markers(mean, "red", "mean")
M.add_markers(a, "blue", "target")
M.generate_spherical_sampled_data()
M.show()
M.show_glyphs()
M.show_pieces()
M.iplot('An arbitrary geodesic')
C) Tangent PCA¶
We've seen how to compute a mean, and how to map our manifold data on a tangent vector space. The next step: analyse the variability in a neighborhood of the mean through a standard, linear statistical analysis.
Exercise 4: Perform PCA in the tangent plane at the mean's location. How many modes do we need to represent the data? Is this a surprise?
Hint: Please use the sklearn.decomposition.PCA module.
## Insert your code here.
# Compute the log-map of the A_k's around the mean
v_k = torch.stack( [ log(mean, a) for a in a_k ] )
# Turn it into an N-by-6 tensor...
v_k = v_k.view(len(v_k),-1)
v_k = v_k.detach().cpu().numpy()
# That we can feed to scikit-learn's PCA module
from sklearn.decomposition import PCA
pca = PCA()
V_k = pca.fit_transform(v_k)
print("Explained variance: ")
print(pca.explained_variance_)
run -i nt_solutions/Kendall_2/exo4
Exercise 5: Display the two principal modes of variation on the Kendall sphere. Is this method accurate enough?
Hint: Use the inverse_transform method of scikit-learn's PCA module.
## Insert your code here.
# We only need the first two features...
pca = PCA(n_components=2)
V_k = pca.fit_transform(v_k)
sig = np.sqrt(pca.explained_variance_)
# Let's retrieve the two directions:
mode_1 = tensor( sig[0]*pca.inverse_transform( [1.,0.] ))
mode_2 = tensor( sig[1]*pca.inverse_transform( [0.,1.] ))
mode_1, mode_2 = mode_1.float().view(3,2), mode_2.float().view(3,2)
# Fancy display
M = KendallTriangles()
M.add_markers(a_k, "green", "A_k", .5)
M.add_markers(mean, "red", "mean")
M.add_markers(exp(mean, mode_1), "blue", "+e_1")
M.add_markers(exp(mean,-mode_1), "blue", "-e_1")
M.add_markers(exp(mean, mode_2), "cyan", "+e_2")
M.add_markers(exp(mean,-mode_2), "cyan", "-e_2")
M.generate_spherical_sampled_data()
M.show()
M.show_glyphs()
M.show_pieces()
M.iplot('The principal modes of variation')
run -i nt_solutions/Kendall_2/exo5
Exercise 6: Display the population as seen through the log-mapping at the mean's location. Is this a faithful representation of the dataset's geometry?
## Insert your code here.
run -i nt_solutions/Kendall_2/exo6
plt.figure(figsize=(10,10))
plt.scatter(V_k[:,0], V_k[:,1], color="green")
plt.scatter( [0.], [0.], color="red", s=125)
plt.scatter( [sig[0],-sig[0]], [0.,0.], color="blue", s=81)
plt.scatter( [0.,0.], [sig[1],-sig[1]], color="cyan", s=81)
midway = plt.Circle((0,0),np.pi/4,color='r', fill=False)
plt.gcf().gca().add_artist(midway)
cut_locus = plt.Circle((0,0), np.pi/2,color='r', fill=False)
plt.gcf().gca().add_artist(cut_locus)
plt.axis("equal")
plt.axis([-np.pi/2,np.pi/2,-np.pi/2,np.pi/2])
plt.title("Exponential chart at the mean location")
plt.show()